The proteins that make up our cells contain an entire universe of data that, when opened, can provide insight into the origins of numerous fundamental organic peculiarities.This data is assembled utilizing a logical method known as “single-cell proteomics,” in which a solitary cell examination is performed to notice the qualities of individual cells at their protein level. Throughout the long term, researchers have been involved in single-cell proteomics in the fields of disease genomics, cell separation, and tissue advancement. Nonetheless, current proteomics methods experience the ill effects of low recovery pace of protein tests, low throughput, and the absence of flexibility.
Luckily, a group of scientists from Japan and the U.S. led by Assistant Professor Takeshi Masuda from Kumamoto University in Japan have tracked down an answer to these issues. In a new report made accessible on July 11th, 2022 and distributed in Analytical Chemistry on July 26th, 2022, the group presented a straightforward yet profoundly effective example of a planning strategy for single-cell proteomics called the “water drop in-oil technique” (WinO). The method utilizes the immiscibility of water with oil or natural dissolvable for its potential benefit to plan protein tests with the least misfortune and expanded possibilities of test recuperation.
“To make single cell-proteomics more effective, we either need to enhance the protein test or ensure none of it is lost during test planning.” Since we didn’t possess the ability to do the previous, it was vital that we decreased retention misfortunes during test planning steps like “example move,” which makes sense for Dr. Masuda. “The WinO strategy lessens test misfortune through adsorption as well as furnishes better throughput when compared with regular techniques.”
“To improve the efficiency of single cell proteomics, we must either amplify the protein sample or ensure that none of it is lost during sample preparation. We needed to reduce absorption losses during sample preparation procedures like sample transfer because we didn’t have the resources to perform the former.”
Assistant Professor Takeshi Masuda from Kumamoto University
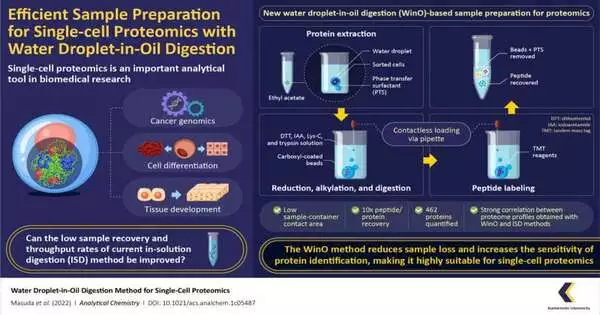
For the WinO cycle, the group initially prearranged an extraction cradle by blending one microliter of water with stage-moving surfactants (which increase the solvency of hydrophobic proteins) and hydrophobic carboxyl-covered nanomagnetic dots. This blend was then dropped into 50 microliters of ethyl acetic acid derivation.
The following stage was protein extraction, which was performed by adding cell drops from the cell sorter to the ethyl acetic acid derivation water bead combo and turning it in a rotator to permit the protein to gather inside the water drop. After the extraction, the example was processed utilizing a protein, Lys-C, and named utilizing a “pair mass tag” reagent. The removed processed named test was then purged and recuperated for single-cell examination and proteomic profiles.
To look at the adequacy of the WinO strategy against regular techniques, the group likewise pre-arranged examples involving the normal in-arrangement processing (ISD) strategy and performed proteomic examination. They discovered that the WinO strategy resulted in a 10-fold increase in protein and peptide recuperation when compared to ISD. This striking improvement was credited to a decreased contact region between the extraction arrangement and the example holder.
To examine the awareness of the two strategies, the group likewise analyzed the proteomic profiles. They noticed a high connection between the proteomic profiles obtained for 100 cells involving WinO and those for 10,000 cells utilizing ISD. Besides, the group effectively measured 462 proteins utilizing WinO, showing that it gave a lot higher throughput and extraction proficiency than regular methods.
The upgraded protein recuperation and ID capacity given by WinO could enable a more critical glance at the protein articulation of disease cells and a superior understanding of the systems’ basic anticancer medication opposition. Further, WinO can be semi-mechanized utilizing a fluid-handling robot, making it reasonable for fast, high-limit handling of tests. “Our exploration could permit researchers to perform proteomics on uncommon and restricted example sums as well as give a clever viewpoint on protein articulation, opening up opportunities for finding new organic peculiarities,” concludes Dr. Masuda.
More information: Takeshi Masuda et al, Water Droplet-in-Oil Digestion Method for Single-Cell Proteomics, Analytical Chemistry (2022). DOI: 10.1021/acs.analchem.1c05487
Journal information: Analytical Chemistry