In order to bridge the gap between basic laboratory science and clinical science and further our understanding of cancer and the development of novel treatments, imaging of tissue specimens is a crucial component of translational research. The ideal application for scientists would allow them to view numerous photographs at once in order to fully study them.
Researchers from the Moffitt Cancer Center explain a novel open-source software tool they created that enables users to view many multiplexed images at once in a paper that was published in the journal Patterns.
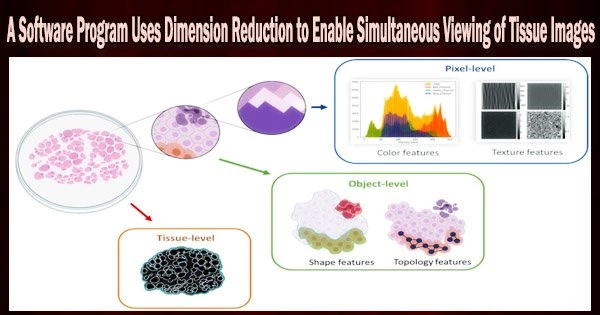
Over the past ten years, there have been major advancements in the methods used to investigate cancer, including fresh methods for examining tissue samples. Today, machines can be designed to stain hundreds of slides at once, or they can be used to concurrently insert up to 1,000 separate tissue sample cores on a single slide and stain them for biomarkers.
The introduction of these methods opens up a wide range of opportunities to produce fresh data and information. Computational modeling and software are required to view and investigate the cancer biomarkers, tissue architecture, and cellular interactions among these samples due to the volume of this information and the complexity of the cancer itself.
As researchers in Moffitt’s Integrated Mathematical Oncology Department (IMO) were working on a project, they realized that the currently available software for image viewing was not amenable to their needs.
We will enhance Mistic to use biologically meaningful regions of interest from the multiplexed image to render the overall image t-SNE. We also have plans to augment Mistic with other visualization software and build a cross-platform viewer plugin to improve the adoption, usability, and functionality of Mistic in the biomedical research community.
Sandy Anderson
“We were interested in understanding the underlying spatial patterns between tumor and immune cells and how the tumors were organized. This required us to compare multiple images simultaneously and we realized there was no software, free or commercial, enabling this,” said Sandhya Prabhakaran, Ph.D., lead author and applied research scientist at Moffitt.
The IMO team made the decision to develop software that would allow them to simultaneously view multiple images and extract data through additional analyses that could be used for a number of purposes, including the discovery of biomarkers and the comprehension of tissue architecture and the spatial organization of various cell types.
Their software, Mistic, abstracts each image to a point in a smaller space using information from multidimensional images and dimensionality reduction techniques called t-distributed stochastic neighbor embedding (t-SNE). Mistic is open-source software that can be used with images from Vectra, CyCIF, t-CyCIF, and CODEX.
The researchers outline the development of Mistic and several potential uses for it in their publication. As an illustration, they showed how the software might be used to examine 92 photos of non-small cell lung cancer patients and determine how biomarkers cluster among patients who had various reactions to treatment.
In another instance, the researchers examined the spatial colocalization and coexpression of immune cell markers in 210 endometrial cancer samples using Mistic in conjunction with statistical analysis.
The team is excited about the potential applications for Mistic and have plans to improve the software.
“We will enhance Mistic to use biologically meaningful regions of interest from the multiplexed image to render the overall image t-SNE. We also have plans to augment Mistic with other visualization software and build a cross-platform viewer plugin to improve the adoption, usability, and functionality of Mistic in the biomedical research community,” said Sandy Anderson, Ph.D., author, and chair of Moffitt’s IMO Department.
In addition to Mistic, the Patterns featured the IMO team in a People of Data article titled “Developing tools for analyzing and viewing multiplexed images.”
The IMO team has the opportunity to introduce themselves, talk about their areas of interest in research, and address opportunities and difficulties specific to imaging in mathematical oncology.