Round ribonucleic acids (circRNAs) are a promising stage for quality articulation. They concentrate as a steady and common ribonucleic corrosive in eukaryotic cells, which emerge from back-joining. In another report currently distributed in Nature Biotechnology, Robert Chen and a group of interdisciplinary scientists at Stanford University, California, U.S., developed a precise way to deal with quickly gathered and tested highlights influencing protein creation in the view of engineered round RNAs. The group boosted interpretation of the circRNA by advancing fine components to execute plan standards to further develop round RNA yield by a few hundred cpm. The results worked with an expanded interpretation of the RNA of interest when contrasted with courier RNA (mRNA) levels, to give a solid interpretation in vivo.
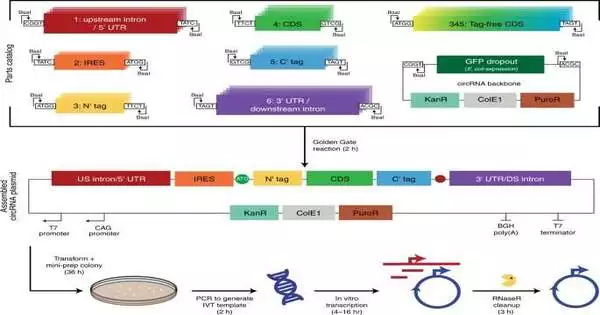
Creating round RNA (circRNA) in the lab
Therapeutics in view of ribonucleic acids range across messenger RNA (mRNA), little meddling RNAs (siRNA) and microRNAs (miRNA) with venture into current medication, including little atoms, biologics, and cell therapeutics. For example, the recently famous mRNA immunizations can be planned in the lab and produced quickly to respond to developing and dire clinical emergencies.Coding RNAs can be circularized into circRNAs to expand the span of protein interpretation in view of RNA atoms that covalently join head-to-tail. Bioengineers have likewise advanced the blend of round, lengthy records into circRNAs. In any case, the key systems of starting interpretation for framing round RNA or courier RNA vary because of the absence of a 7-methylguanylate (M7G) cap on the round RNAs. Thus, analysts need to completely inspect the standards of round RNA interpretation to assemble better treatments and possibly outperform the translational limits of mRNA. To inspect this viewpoint, the group fostered a secluded high-throughput stage to fabricate and test engineered round RNAs for upgraded interpretation and further developed protein yields.
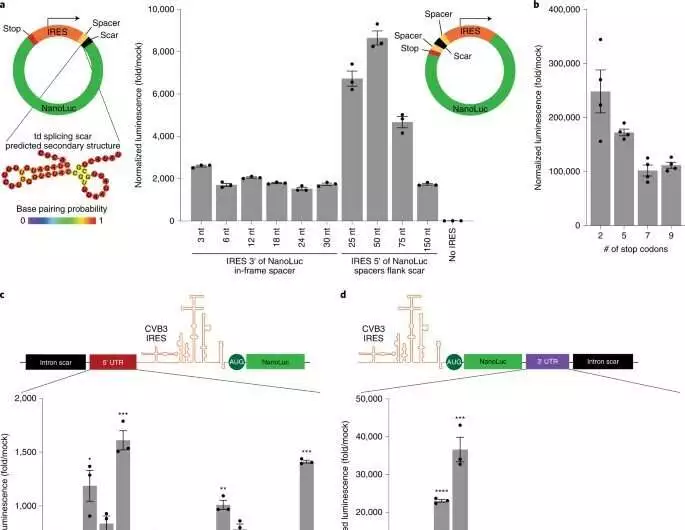
Enhancement of RNA non-coding components empowers more grounded circRNA interpretation. (a) NanoLuc activity after HeLa cells were transfected with circRNAs containing either a 3′ or a 5′ IRES and spacer groupings of varying lengths.At the point when the IRES is 3′ to the NanoLuc columnist, interpretation through the td joining scar is undeniable. The anticipated optional design of this scar is shown. The NanoLuc action was standardized to constitutive firefly luciferase action from a similar example and afterward isolated by values from mock transfection. For n = 3 organic repeats, the information is mean s.e.m.(b) NanoLuc action at 24 hours after transfection of HeLa cells with circRNAs containing the shown number of stop codons. The NanoLuc action was standardized to constitutive firefly luciferase action from a similar example and afterward isolated by values from mock transfection. For n = 4 organic repeats, the information is mean s.e.m.(c) NanoLuc activity after transfection of HeLa cells with circRNAs with various 5′ spacer groupings.The NanoLuc action was standardized to constitutive firefly luciferase action from a similar example and afterward isolated by values from mock transfection. For n = 3 organic repeats, the information is mean s.e.m.An unpaired two-sided t-test yielded ***P 0.001 when compared to an irregular 50-nt spacer grouping.(d) NanoLuc activity after transfection of HeLa cells with circRNAs with various 3′ UTR groupings.The NanoLuc action was standardized to constitutive firefly luciferase action from a similar example and afterward isolated by values from mock transfection. For n = 3 organic repeats, the information is mean s.e.m.***P = 0.0012 and ****P < 0.0001 by unpaired two-sided t-test contrasted with an irregular 50-nt spacer grouping. BR stands for limiting region; MR stands for minor region; and PR stands for protected region.Nature Biotechnology (2022) is credited.DOI: 10.1038/s41587-022-01393-0
A measured circRNA gathering stage.
The researchers fostered a secluded cloning stage made of a bunch of parts viable with Golden Gate and Gibson cloning to permit higher-throughput testing of circRNAs. Using the stage, they decided what explicit parts of the round RNA configuration were meant for its interpretation. For example, the group has recently shown how round RNA sets off safe reactions that can be tried not to in vivo by altering the atoms with m6A. In any case, analysts should grasp the effect of this step on round RNA interpretation in any case. To address this, Chen and the group utilized their cloning stage and integrated m6A. When contrasted with unmodified circRNAs, those containing 5% m6A showed equivalent interpretation after transfection or electroporation in vitro. The researchers hence tentatively checked the effect of the change on round RNA security.
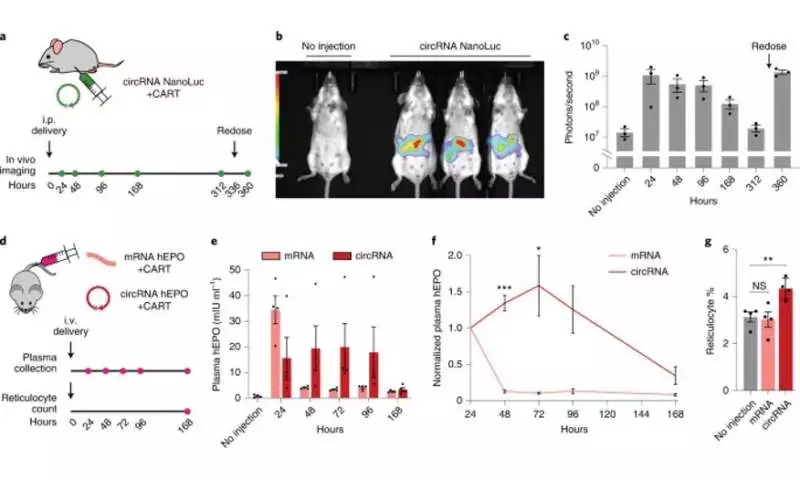
Revealing the elements of circRNA for solid interpretation results
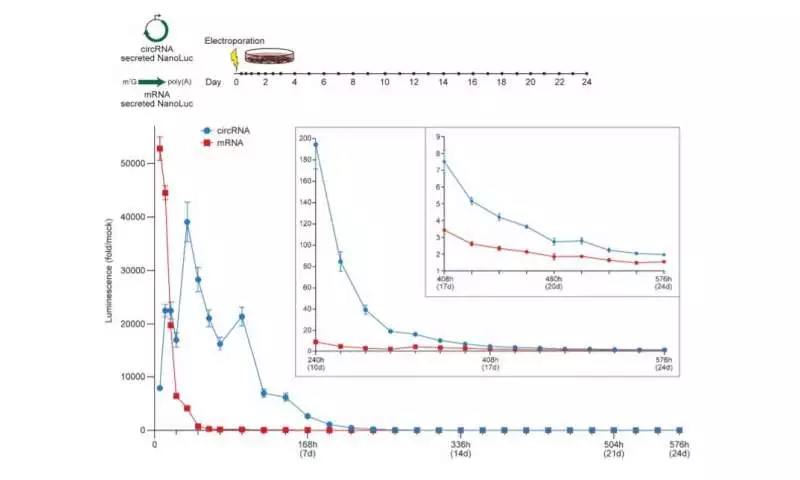
To reveal the standards’ basic circRNA vector geography vital for solid interpretation, the analysts started blending circRNAs to create variations with peptides encoded by the cycle. In view of the results, the group showed that expanding the spacer-length was not useful for interpretation. Then, they demonstrated the way that the 5′ and 3′ untranslated areas could further develop circRNA interpretation. The scientists also led a progression of tests to look at circRNA enhancement and afterward analyzed them in a solitary trial. They showed how the progressions logically expanded the outflow of circRNA without compromising the RNA yield or proficiency of circularization. They likewise showed how the energy of circRNA and mRNA interpretation altogether varied, where circRNA required over 24-hours to arrive at its greatest interpretation length, far surpassing the interpretation term of mRNA. They then joined the series of circRNA enhancements to test their demeanor in vivo. To convey the RNAs, the group formed them with charge-changing releasable carriers (CARTs) or cationic atoms that intervene in mRNA articulation in mouse models. The results showed how the designed circRNAs could be communicated at quality like changed RNAs in vivo, yet with a more prominent span.
Viewpoint
Along these lines, Robert Chen and partners showed how RNA circularization can possibly change RNA-based meds by expanding the strength of somewhat profoundly transient atoms. Given the key distinctions between the systems of circRNA and mRNA interpretation, the current information on boosting mRNA interpretation couldn’t be guaranteed to mean circRNAs. To work with this review, the group made a circRNA measured cloning stage to test various succession varieties and enhancements of numerous boundaries. Utilizing the stage, they recognized a few ways to deal with further developing protein interpretation from circRNAs, with applications to extensively designing RNAs, creating more proteins than mRNAs in vitro, and showing more prominent strength of their interpretation in vivo and in vitro. The researchers efficiently analyzed the components managing circRNA interpretation to then upgrade the areas of interest for expanded circRNA protein yields for solid protein creation in vivo.
More information: Robert Chen et al, Engineering circular RNA for enhanced protein production, Nature Biotechnology (2022). DOI: 10.1038/s41587-022-01393-0
Chang-you Chen et al, Initiation of Protein Synthesis by the Eukaryotic Translational Apparatus on Circular RNAs, Science (2006). DOI: 10.1126/science.7536344