Understanding the reason why and how chemotherapy opposition happens is a significant stage toward upgrading therapies for malignant growth. A group of researchers including Markus Seeliger, Ph.D., of the Stony Brook Cancer Center and Renaissance School of Medicine at Stony Brook University, accept they have found another interaction through which drug opposition occurs. They are utilizing a virtual experience model that is assisting them with seeing precisely the way in which particles collaborate with the disease drug Imatinib (known as Gleevec) in the chemotherapy safe cycle. Imatinib treats persistent myeloid leukemia (CML) profoundly actually, yet many late stage patients experience drug obstruction, which delivers the medication negligibly viable at that stage.
The examination is featured in a paper distributed in Angewandte Chemie and expands upon past exploration point by point in 2021 in PNAS.
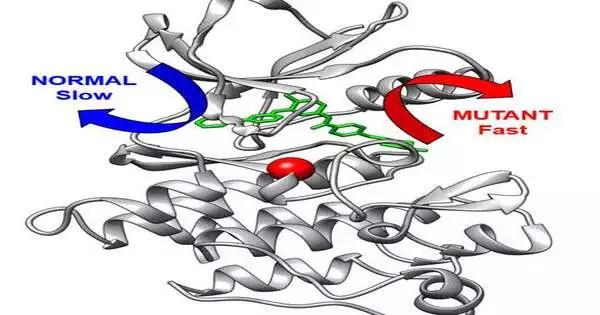
Imatinib restrains the BCR-Abl protein kinase, an excessively dynamic cell flagging apparatus in CML. In the PNAS study, specialists showed that varieties in the structure plan of the kinase can make it harder for Imatinib to tie to the kinase and furthermore accelerate drug discharge from the kinase. In the Angewandte Chemie paper, the exploration group took the computational philosophy — created by co-creator Pratyush Tiwary from the University of Maryland — that empowered them to concentrate on the extremely sluggish arrival of Imatinib from the kinase.
“This strategy in and of itself is a significant technological success that broadens computational capabilities for drug resistance research and, more crucially, enabled us to anticipate how quickly healthy and mutant proteins would release this drug,” the researchers write.
Seeliger, Associate Professor in the Department of Pharmacological Sciences.
“This strategy in itself is a significant specialized accomplishment that expands computational capacities for drug obstruction research, and critically prompted us having the option to foresee how quickly solid and freak proteins would deliver this medication,” says Seeliger, Associate Professor in the Department of Pharmacological Sciences. “Interestingly, we could see the arrival of a medication from a protein in such detail and precision. Besides, we could show that the transformation changes on a very basic level inside the leave course of the medication from the protein.
“This is significant since the speed of the medication delivery might be similarly as significant for the helpful impact of a medication as how firmly a medication ties to the protein.”
Seeliger further makes sense of that the technique could give an establishment to grasping the sub-atomic systems behind chemotherapy obstruction.
All the more extensively, the ramifications of what they found are that on the off chance that researchers can comprehend how medications are let out of their proteins, they might have the option to configuration drugs with a more slow delivery and higher helpful effect. Also, assuming quick medication delivery could cause drug obstruction, and clinicians can show this is going on, they might have the option to re-enact the medication adequacy by requesting that the patient take the medication all the more often.
The basis for the change testing by means of the computational strategy was illustrated in the PNAS paper. Seeliger and partners tried how imatinib ties to transformations in patients with imatinib-safe CML. They found that most of changes promptly tie to imatinib, so that suggested the conversation starter exactly how do these transformations cause obstruction in patients? The scientists then, at that point, recognized a few freaks which bound imatinib promptly however they discharge the medication a lot quicker.
Subsequent to recognizing these freaks with a quicker drug discharge, the group utilized atomic attractive reverberation (NMR) and sub-atomic elements to connect the protein to sedate disassociation — basic the significance of medication disassociation energy for drug viability. This empowered them to personality a clever component of imatinib obstruction.
The work bringing about the paper distributed in PNAS involved the cooperative endeavors of Seeliger and his associates at Stony Brook, and specialists at Memorial Sloan Kettering Cancer Center and at Goethe University of Frankfurt, Germany.
Research that brought about the later paper was driven by Tiwary and partners at the University of Maryland, in a joint effort with Seeliger and researchers at the Broad Institute at MIT and Harvard University.
More information: Mrinal Shekhar et al, Protein Flexibility and Dissociation Pathway Differentiation Can Explain Onset of Resistance Mutations in Kinases, Angewandte Chemie International Edition (2022). DOI: 10.1002/anie.202200983