UVA Wellbeing Disease Center scientists have fostered a calculation that will further develop malignant growth care by rapidly and effectively recognizing patients who will profit from strong disease drugs called kinase inhibitors. The calculation might have other analytic advantages for patients too.
Kinase inhibitors are the most well-known disease drugs endorsed by the government’s Food and Medication Organization. They can be immensely viable for the right patients, yet they don’t work for everybody. UVA’s calculation offers a new and better method for pinpointing patients who will benefit—a significant step in the right direction in medication accuracy customized to the person.
“We are truly amped up for this calculation, which performs better compared to existing methodologies with fewer necessities and suspicions, making it more relevant to understanding a disease state from a solitary preview of the growth,” said scientist Kristen M. Naegle of UVA’s Branch of Biomedical Designing, a program of UVA’s schools of medication and design working together. “Joining this methodology with existing biomarkers for disease finding might assist us with bettering designer treatments, planning new mixes of treatments, expecting reaction to treatment, and configuring better clinical preliminaries.”
“We are really excited about this algorithm, which performs better than existing approaches with fewer requirements and assumptions—making it more applicable to understanding a cancer state from a single snapshot of the tumor,”
Kristen M. Naegle of UVA’s Department of Biomedical Engineering,
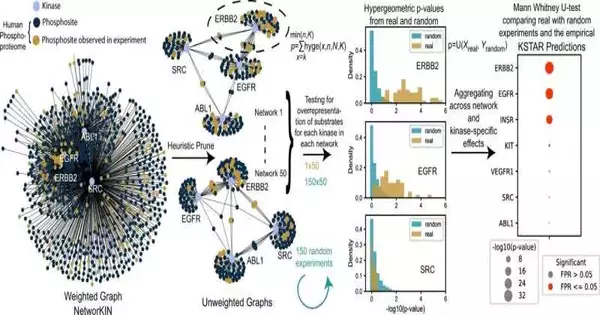
KSTAR for better disease care.
Naegle and colleagues set out to test the limits of existing techniques to recognize patients who might profit from kinase inhibitors. The majority of these strategies require hard-to-get and, at times, problematic data measuring “phosphorylation locales” inside cells. UVA’s new methodology, nonetheless, needn’t bother with this intricate estimation. All things considered, it can foresee key data in light of other accessible information.
This permits the calculation to create a particular “KSTAR score” for individual proteins called kinases. Specialists can utilize these scores to figure out which patients will respond to kinase inhibitors, helping guide the best treatment decisions. Kinase inhibitors are broadly utilized for specific kinds of blood, bone, and cellular breakdowns in the lungs, among others. These medications are frequently used in conjunction with other therapies such as chemotherapy, radiation, or medical procedure.
In testing their new calculation, Naegle and her partners found that it worked dependably across various tissue types, suggesting it might be helpful for some sorts of disease.
As an additional advantage, KSTAR can act as an indicative device, the UVA scientists report. During their tests, KSTAR had the option to verify that a patient with bosom disease was not HER2 positive, as specialists had recently accepted. HER2-positive bosom diseases can profit from HER2-designated kinase restraint, yet HER2-negative growths will not, and HER2-hindrance accompanies unexpected confusions.
Moreover, the scientists see that around 20% of patients recognized as HER2-negative by current clinical methodologies really have HER2 marks that suggest they could profit from HER2-designated treatment—a treatment not presently proposed to them. That kind of data can be priceless as specialists and patients examine treatment choices.
“We are teaming up and working with groups of scientists across a scope of tumors to lay out when and how KSTAR can assist with recognizing patient reaction to treatment,” Naegle said. “We trust that we will actually want to assist better with recognizing the right treatment for the ideal patient brilliantly for improved results for patients.”
Naegle and her partners have made their new calculation openly accessible. They have published their discoveries in Nature Correspondences.
More information: Sam Crowl et al, KSTAR: An algorithm to predict patient-specific kinase activities from phosphoproteomic data, Nature Communications (2022). DOI: 10.1038/s41467-022-32017-5
Journal information: Nature Communications