An approach has been developed to create artificial allosteric sites in protein complexes, whereby activity at the distal active site is controlled by binding an effector molecule, according to a recently published research paper. This leading-edge research holds huge promise for a large number of uses in modern, organic, clinical, and rural fields. The work was distributed in Nature Science.
Subunits (constituent proteins in the protein complex) collaborate to perform coordinated functions in protein complexes like hemoglobin and molecular motors. This organization is empowered by the allosteric system.
Since its inception in the 1960s, the concept of allosteric effect—the regulation of function at an active site in one subunit by the binding of an effector molecule to an allosteric site in another subunit—has remained one of the most pressing issues in biochemistry.
“Artificial allosteric sites introduced into protein complexes have the potential to reveal fundamental allostery principles and provide tools for synthetic biology,”
Nobuyasu Koga, a professor at the Osaka University.
An examination group has fostered a technique for planning counterfeit allosteric destinations into protein edifices to control the purposeful capability of a protein complex. “The making of fake allosteric destinations into protein edifices can possibly uncover crucial standards for allostery and act as devices for manufactured science,” said Nobuyasu Koga, a teacher at Osaka College.
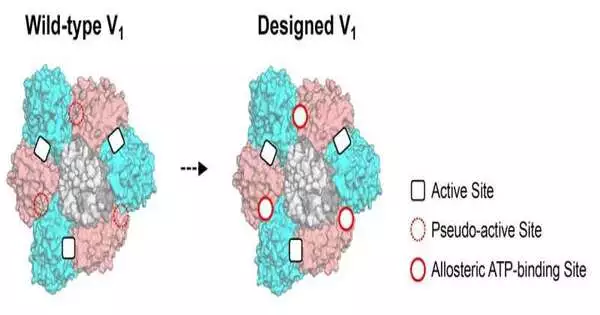
The exploration group conjectured that allosteric locales in protein edifices can be made by reestablishing lost elements of the pseudo-dynamic destinations, which are anticipated to have been lost during development. Different protein edifices incorporate subunits that have pseudo-dynamic destinations.
Pseudo-active sites have been shown to have an allosteric link to active sites in other subunits. For instance, a pseudo-dynamic site in a subunit that has lost ATPase movement yet at the same time shows ATP-restricting capacity enacts one more subunit’s dynamic site after restricting to ATP. At the cell level, ATP is the wellspring of energy. The enzyme’s ability to break down ATP is referred to as ATPase. These studies back up the idea that protein complexes can be made up of different allosteric sites by engineering pseudo-active sites.
The V1-ATPase’s pseudo-active site in the B-subunit of a rotary molecular motor, which had lost its ability to bind ATP, was reactivated through the use of computational design by the research team. In the first place, the restricting skill of the planned site was tentatively uncovered by X-beam crystallography. “The X-beam structure showed the limiting site is effectively planned and incorporated into the regular protein to have a capability. An associate professor at the Institute of Materials Structure Science named Mikio Tanabe shared, “I was amazed at the utility and high performance of protein design technology.”
Following that, their single-molecule experiments and X-ray crystallography analyses revealed that the rotation rate can be adjusted by altering the ATP’s binding affinity and that ATP binding to the designed allosteric site increases this V1’s activity in comparison to the wildtype. The rotary motor’s cooperation was created by the team.
In addition, when compared to the wildtype, the team’s designed allostery accelerated rather than slowed the rotation. To the best of our knowledge, this is the first time that protein engineering has been used to accelerate a rotary molecular motor According to Ryota Iino, a professor at the National Institutes of Health, “This is an exciting result in the field.”
Planned allostery helped revolutionize the planned V1. Credit: Pseudo-active sites like NINS and IMS are common in nature and could be used to program allosteric control over coordinated protein complex functions. Furthermore, the protein plan strategy would empower not only to reestablish the lost capability but also to restrict destinations for different ligands.
“In principle, we can create allosteric sites in a variety of protein complexes using our method. Our next stage is to make allosteric control for an assortment of protein buildings using our procedure. Also, we will attempt to plan novel capabilities into pseudo-dynamic locales. One of our objectives is to misleadingly control the purposeful elements of any protein edifice and to uncover the overall component of allostery. According to Takahiro Kosugi, an assistant professor at the National Institutes of Health, “We hope allosteric control over concerted functions of protein complexes will open up new avenues in industrial applications of enzymes in biological, medical, and agricultural fields.”
More information: Design of allosteric sites into rotary motor V1-ATPase by restoring lost function of pseudo-active sites, Nature Chemistry (2023). DOI: 10.1038/s41557-023-01256-4