Scientists from A*STAR’s Genome Organization of Singapore (GIS) have tracked down an imaginative way to deal with sequencing single-cell ribonucleic corrosive (RNA) to concentrate on the jobs of RNA structure in individual cells. As RNA shapes in individual cells can change and drive various capabilities, concentrating on the aggregate RNA shapes in numerous cells might neglect the singular fluctuation in both RNA construction and capability.
Named Sc-Game for Single Cell Design Testing of RNA Records, this new methodology can distinguish biomarkers urgent for human turn of events and illness in view of shape as opposed to grouping. While single-cell quality articulation examination has shown the degree of variety that is available in apparently comparative individual cells, it is unclear whether different components could likewise impact cell destiny. Through this methodology, scientists can now utilize RNA structure as an extra degree of data to recognize cell types being developed and illnesses.
The paper, “RNA structure profiling at the single-cell goal uncovers new determinants of cell personality,” was distributed in Nature Strategies.
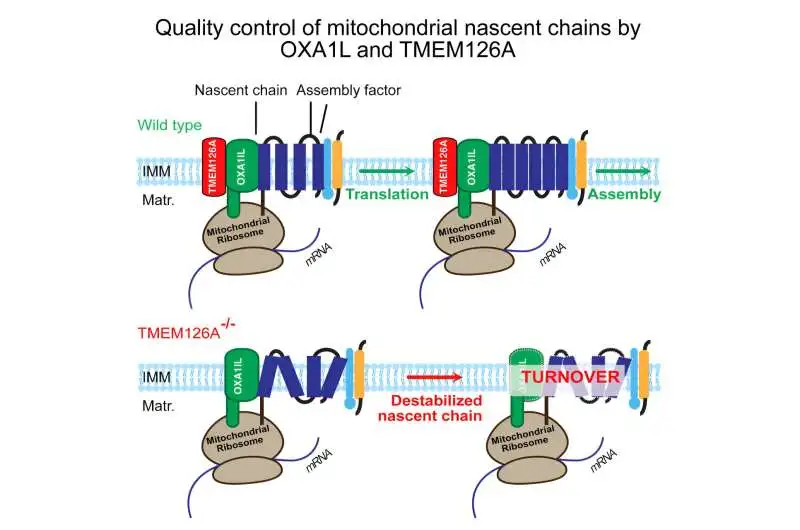
“This discovery is a critical step toward developing fresh treatment strategies for afflicted individuals. Finding the sources of some disorders may be made easier by comprehending the interactions between proteins in mitochondria. Knowing which processes in specific disorders are malfunctioning or what is lacking from the cell allows us to ‘repair’ this problem through treatment.”
Prof. Dr. Peter Rehling, Director of the Department of Cellular Biochemistry at the University Medical Center Göttingen (UMG).
In the previous ten years, understanding various parts in individual cells through different single-cell sequencing advancements has advanced huge amounts at a time. As specialists dive further into the layers of individual cells, foreseeing cell directions turns out to be progressively exact.
Driven by A*STAR’s GIS Representative Leader Chief and Head Examiner, Dr. Wan Yue, and A*STAR’s GIS individual, Dr. Jiaxu Wang, in a joint effort with Dr. Roland Huber, Head Specialist, A*STAR’s Bioinformatics Foundation (BII), this study denotes the primary investigation of RNA structure at a solitary cell level, explicitly focusing on uncommon cell types and RNA structure heterogeneity. Before, while concentrating on the design of RNA in enormous amounts, specialists expected a huge number of cells to start.
Such a methodology prompted a nonexclusive outline, consolidating every one of the shapes from every one of the cells, which made it hard to see the particular distinctions in RNA structure among individual cells. Presently, by performing RNA structure examinations in single cells, specialists can more readily value the data present in every cell and recognize when the cells could turn out badly.
The investigation of RNA structure in single cells opens ways to recognizing structure biomarkers in individual cells that can be dysregulated in illness, adding an extra layer of data to existing single-cell data. This RNA construction could act as a novel biomarker or drug focus for illnesses in which the RNA levels don’t change.
It would empower specialists to distinguish cell types in view of RNA structure, notwithstanding RNA articulation, and better comprehend how RNA infections can overlap in individual cells. Presently, the group is streamlining approaches for single-cell RNA structure sequencing and applying this method to deal with more mind-boggling cell formative cycles and disease.
Graphical abstract. Credit: Molecular Cell (2024). DOI: 10.1016/j.molcel.2023.12.013
Dr. Wan Yue said, “Not at all like conventional techniques that concentrate on a large number of cells immediately, this advancement empowers looking at RNA structures in every cell exclusively, revealing special examples and biomarkers, and enhancing how we might interpret cell destiny. Despite the coronavirus pandemic brought about by an RNA infection, understanding how RNA folds inside cells becomes principal.”
“This study not only reveals insight into the essential elements of RNA but additionally makes ready for remarkable experiences into the capability of RNA as a distinct advantage in biomedical sciences.”
The discoveries in this most recent paper are likewise one of a kind, contrasted with before concentrates, because of their capacity to decrease a huge number of cells to a solitary cell level for RNA structure sequencing.
Acting Leader Chief at A*STAR’s GIS, Teacher Liu Jian Jun, said, “In the quick-developing scene of RNA research, the work led at GIS underlines our obligation to unravel the intricacies of RNA. The uplifted attention to RNA’s job, particularly with regards to the coronavirus pandemic, adds to better pandemic readiness and offers new experiences for likely medicines against RNA infections.”
More information: Sabine Poerschke et al, Identification of TMEM126A as OXA1L-interacting protein reveals cotranslational quality control in mitochondria, Molecular Cell (2024). DOI: 10.1016/j.molcel.2023.12.013