A group of researchers at the University of Oxford have worked with different strategies at the precious stone light source to settle the construction of the flu replication hardware and determine how it interfaces with cell proteins. This exploration promotes how we might interpret flu replication and how the infection adjusts to various hosts.
Past causing occasional influenza, flu can become pandemic when it bounces from creatures to people. By investigating the replication pattern of the infection, analysts are sorting out how flu commandeers human and creature cells for its replication.
A new survey in Patterns in Microbial Science frames the discoveries created utilizing X-beam crystallography and little point X-beam dispersing (SAXS) at the synchrotron and cryo-electron microscopy (cryo-EM) at Precious Stone’s electron bio-imaging center (eBIC).
“These investigations aid in the identification and validation of drug development targets. We hope that the new insights gained into the workings of the influenza virus transcription machinery as a result of Diamond’s innovations may eventually lead to innovative antivirals targeting the influenza polymerase.”
Professor Ervin Fodor, University of Oxford,
These primary experiences have uncovered new potential medication focuses for the improvement of novel antiviral medications to hinder flu infection replication.
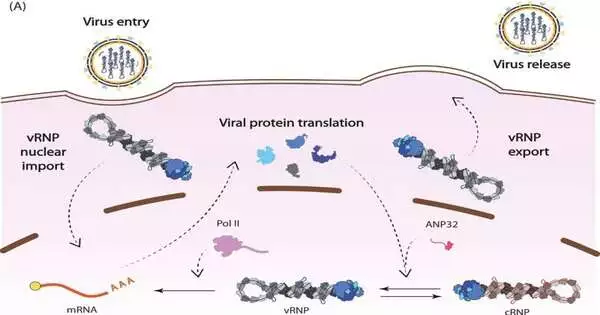
The flu infection stores its qualities in RNA, and the infection orchestrates its own RNA polymerase to duplicate its genome. This viral polymerase has different capabilities notwithstanding replication, which cooperative examination across Jewel has assisted in clarifying.
These examinations show that the polymerase manages the planning of transcription—tthe most vital phase in protein combination—aand replication, which can start once popular proteins have been delivered. The discoveries uncover how the polymerase communicates with a cell protein, ANP32A, and appropriates it to protect viral RNA from being located by the insusceptible framework.
The present circling flu Infections are believed to be the developmental descendants of the infection that caused the 1918–1919 worldwide pandemic, which was responsible for somewhere in the range of 50 and 100 million deaths around the world. Flu infections are typically limited to contaminating one sort of creature, like birds, and require explicit transformations to leap to an alternate creature, similar to people.
The 1918 flu infection is remembered to have hopped from waterfowl into people and is viewed as the “organizer infection” that has contributed viral genome portions for all resulting scourges and pandemic strains. In a review distributed recently, the gathering decided the designs of the polymerase from the 1918 pandemic flu infection and distinguished destinations on the outer layer of the polymerase that are delicate to inhibition 1. This thus can help distinguish and approve focuses for drug revelation.
This examination is pivotal to understanding how ANP32A somewhat represents the host-hopping obstruction. ANP32A contrasts incredibly between people and birds, compelling creatures and avian flu infections to develop and turn out to be less indistinguishable.
Primary science research at Jewel gives experiences into the pandemic capability of various kinds of influenza. By concentrating on what locales of the viral polymerase cooperate with ANP32A, the specialists discovered that a transformation in avian flu’s polymerase might permit it to connect with human ANP32A, allowing that type of bird influenza to hop into human hosts 2.
The underlying portrayal of enormous protein edifices is a test, and the flu replication complex was no exemption. X-beam crystallography at beamlines I03 and I24 was utilized to determine the design of the viral polymerase in close nuclear detail, uncovering that the single polymerases pair together to shape dimers. To supplement the gem design of the dimers, a primary method known as SAXS was used in an arrangement at beamline B21 to show the significance of dimer development to the capability of the polymerases.
The specialists suggested that solitary RNA polymerases complete record from the get-go in disease and change to replication later just when they couple together as dimers, following the creation of extra copies of the polymerase3.
To develop this underlying work further, the examination group performed cryo-EM at eBIC. Teacher Jonathan Grimes at the College of Oxford makes sense of the fact that “cryo-EM has permitted us to start to take a gander at exceptionally fascinating protein edifices that we would find difficult to develop precious stones of in the lab.”
“Jewel democratizes science.” The fact that these strategies exist in one spot and are accessible to mainstream researchers is a massively significant asset. These a-list state-of-the-art offices are unreservedly accessible to researchers from colleges and organizations across the UK and EU with intriguing and significant natural inquiries.
Comparing the Patterns in Microbial Science Audit, Teacher Ervin Fodor, College of Oxford, reasons that “these examinations assist us with distinguishing and approving foci for drug revelation.” “We are trusting that the new bits of knowledge created about the functions of the flu infection record apparatus utilizing the advancements at Precious Stone will ultimately prompt novel antivirals focusing on the flu polymerase.”
More information: Zihan Zhu et al, A structural understanding of influenza virus genome replication, Trends in Microbiology (2022). DOI: 10.1016/j.tim.2022.09.015