Food-borne transmission of critical-priority antimicrobial-resistant enterobacterales is a public health issue. Wild-caught seafood is widely consumed around the world, but exposure to land-based pollution can increase the likelihood of contamination by clinically relevant antimicrobial-resistant bacteria. As part of the Grand Challenges Explorations: New Approaches to Characterizing the Global Burden of Antimicrobial Resistance Program, we conducted genomic surveillance and cell culture-based virulence investigations on WHO critical priority Enterobacterales isolated from marine bivalves collected along South America’s Atlantic Coast. Klebsiella pneumoniae and Escherichia coli isolates with broad-spectrum cephalosporin resistance were obtained from eight different geographical areas. These strains carried the blaCTX-M or blaCMY genes. The majority of the genomes examined confirmed the convergence of a large virulome and resistome (i.e., antimicrobials, heavy metals, biocides, and pesticide resistance).
We isolated isolates from the international high-risk clones K and L. pneumoniae ST307 and E. pneumoniaeIn vitro tests revealed that they have a high virulence potential.Some strains produced thermostable and thermostable toxins, and all of them produced biofilm. These findings suggest that resistance and virulence genes are abundant in marine habitats, which may facilitate horizontal gene transfer and the spread of these traits to other bacterial species.
Introduction
The fast and widespread spread of critical-priority antimicrobial-resistant Enterobacterales is a public health issue that necessitates mitigation strategies and strengthening through genomic epidemiological surveillance. Given its epidemiological importance, there has been an increase in reports of critical-priority bacteria in aquatic environments2, including coastal waters3,4,5, implying that polluted marine environments may serve as potential reservoirs for waterborne pathogens.
Consumption of seafood has increased dramatically in recent decades. Despite the fact that domestic aquaculture production has shown a linear trend of growth, wild-caught seafood is still widely consumed globally (7, 8, 9). Marine bivalves have a wide geographic distribution in this context and have long been regarded as an alternative food protein source for coastal populations worldwide10.Because of their activity in filtering particles floating in the surrounding water, they have also been utilized as marine sentinels and examined as potential ecological bioindicators to detect environmental damage connected to water pollution11.
In recent studies, human gastroenteritis outbreaks have been linked to the eating of infected bivalves (12,13,14). In this context, norovirus and Vibrio spp. The scientific community’s interest in infections has piqued due to their pathogenicity and virulence13,14,15. Although some studies have postulated that the discovery of multidrug-resistant (MDR) bacteria in marine bivalves could pose a risk to seafood consumers, 12–16, 17–there is little scientific data to support this claim.
The rise of critical priority enterobacterales producing CTX-M-type and CMY-type-lactamases, as well as thermolabile and thermostable toxins, in wild-caught marine bivalves off the Southeast coast of Brazil, South America’s largest and most populated country, is described here. We conducted a thorough examination utilizing WGS and in vitro studies to assess these bacteria’s ability to infect human cells, revealing a new and hidden threat to seafood consumers.
Results
Multidrug-resistant clinically relevant bacteria are found in edible marine bivalves.
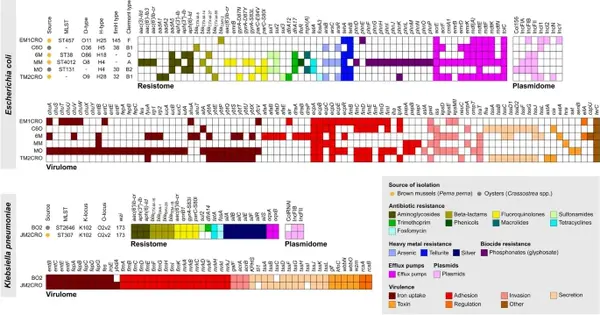
Eight ceftriaxone-resistant Enterobacterales, comprising E. coli (n = 6) and K. pneumoniae (n = 2) isolates, were isolated from edible marine bivalves collected at various locations along Brazil’s southeast coast (Fig. 1 and Table 1). Brown mussels had four E. coli and one K. pneumoniae, while oysters contained two E. coli and one K. pneumoniae. All isolates had an MDR resistance profile but were still vulnerable to carbapenems and colistin (Fig. 2). Seven isolates demonstrated an ESBL characteristic, with cefotaxime MICs greater than 32 g/mL. In addition, one E. coli strain (EM1CRO) was cefoxitin-resistant (MIC > 32 g/mL).
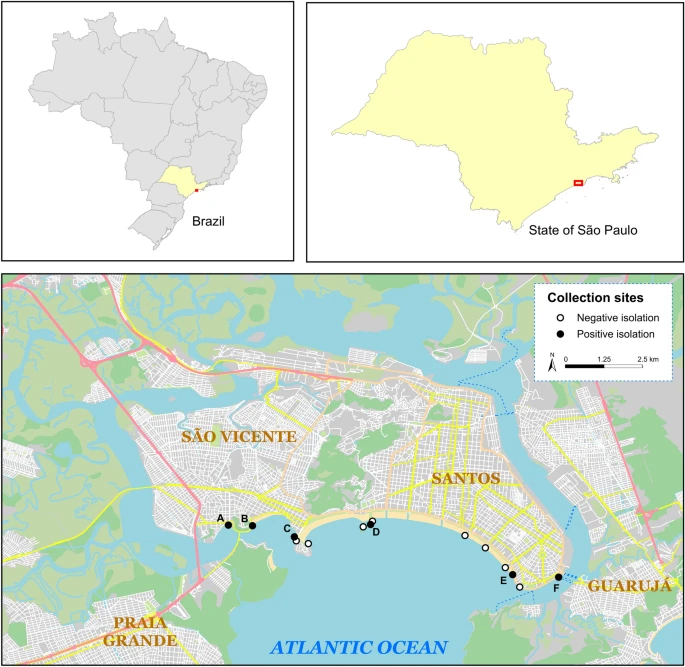
A map depicting 14 separate collection sites along the coasts of So Vicente and Santos, both cities in Southeast Brazil. Priority number one Enterobacterales strains were recovered from bivalves at 6 of these sites (indicated by black markers): (A)—E. coli TM2CRO [23.972588 S, 46.39192 W]; (B)—K. pneumoniae JM2CRO [23.972765 S, 46.384877 W]; (C)—E. coli MM and E. coli MO [23.976040 S, 46.372580 W]; (D)—E. coli EM1CRO [23.972388 S, 46.35024 The map was created using public domain shapefiles provided by the Brazilian Institute of Geography and Statistics (https://www.ibge.gov.br/) and OpenStreetMap vector layers (OpenStreetMap contributors), both of which are released under the Open Data Commons Open Database License. (https://www.openstreetmap.org/copyright). OpenStreetMap vector layers were downloaded from QGIS v3.22.4 (https://qgis.org) with the OSMDownloader plugin v1.0.3 (https://github.com/lcoandrade/OSMDownloader). ESRI ArcMapTM v10.7 was used to create the final layout.
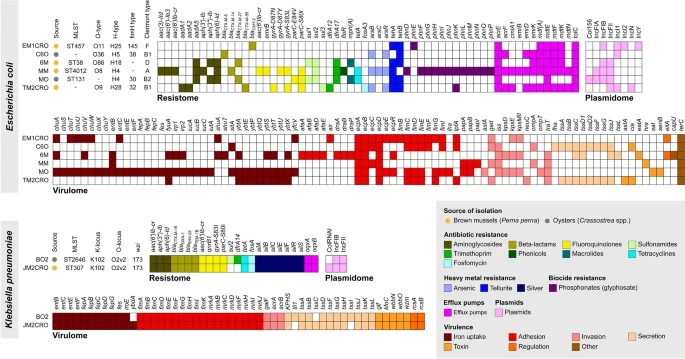
In seafood bacterial isolates, genomic investigations reveal the convergence of a wide virulome and a resistome.
Resistome data from E. EM1CRO, MO, and 6M have previously been described in an epidemic context (18, 19). In the current work, we performed a comprehensive genomic analysis that includes new E. E. coli C60, MM, and TM2CRO strains, as well as K. pneumoniae strains BO2 and JM2CROFigure 2 depicts the main findings of the WGS analysis. The convergence of a broad virulome and resistome (antimicrobials, heavy metals, biocides, and pesticide resistance genes) was confirmed in the majority of the genomes studied. Except for E. Among E. coli EM1CRO, all strains tested positive for CTX-M variations, including blaCTX-M-8 (C6O), blaCTX-M-14 (MM), blaCTX-M-15 (BO2 and JM2CRO), blaCTX-M-27 (MO and 6M), and blaCTX-M-55 (TM2CRO). blaCMY-2, which was positive for the plasmid-mediated AmpC (pAmpC) gene. Unfortunately, because we employed short read sequencing techniques, we were unable to completely assemble the plasmids to verify all genes and associated plasmid replicons. However, using the mlplasmids program (https://sarredondo.shinyapps.io/mlplasmids/), the genes identified in plasmids for E. A number of coli strains could be predicted (EM1CRO: mdfA; 6M: blaCTX-M-27; MO: aadA5, sul1, dfrA17, mphA; and TM2CRO: blaCTX-M-55).
Most strains have a diverse virulence repertoire, with lineages from phylogroups B2 (MO) and D (6M) having a larger virulome than those from phylogroups A (MM), B1 (C6O and TM2CRO), and F. (EM1CRO). The MDR strains belonged to various STs and, concerningly, an E. coli strain of ST131 (MO) and a K. A pneumoniae strain of ST307 (JM2CRO) was found. Overall, E. coli strains exhibited a significant number of genes involved with host adhesion (i.e., iha, lpf, pap, ecp, fim, and afa/dr fimbriae), host immune system resistance (i.e., kps and neuC capsular polysaccharides; tra and iss complement and serum resistance), and host damage (i.e., senB and astA (i.e., chu, iuc, ent, irp, and ybt). E.g.coli strains had a variety of serotypes and fimbriae types, including O11: H25, O36: H5, O86: H18, O8: H4, and O9: H28; fimH145, fimH38, fimH30, and fimH32. Pneumoniae (K.Pneumoniae) strains also revealed genes involved with host adhesion (i.e., fim operon), host immune system resistance (i.e., kfoC complement and serum resistance), host damage, and iron/hemin absorption (i.e., iro and ent). The K-locus KL102/wzi173 was also found in BO2 and JM2CRO K. Pneumoniae strains The assembly of the genome revealed that IncF-type plasmids were the most common among the strains. Because of the constraints of short-read sequencing, only the kind of plasmid (IncFII) linked with the blaCTX-M gene in the K. A Pneumoniae BO2 strain could be determined.
Enterobacterales isolated from edible marine bivalves exhibits virulent activity.
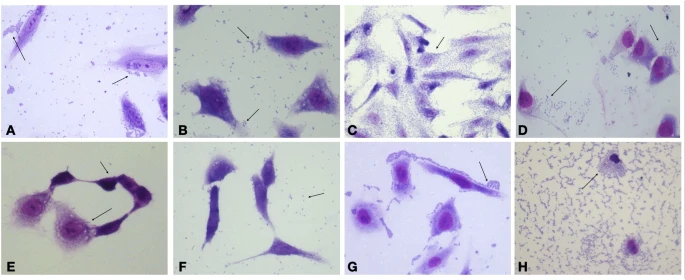
Bacterial isolates were tested to see if they could colonize and harm human epithelial cells. In this regard, all E. coli isolates were capable of adhering to HeLa cells after 6 h of incubation (Fig. 3), despite the fact that their adherence patterns differed, with EM1CRO and C6O strains exhibiting aggregative adherence, 6M and TM2CRO strains adhering diffusely to the cells, and MM and MO exhibiting an undefined adherence pattern. The two K. pneumoniae strains also showed aggregative adhesion on the surface of the epithelial cells as well as on the glass slide. Furthermore, after 6 hours of incubation, E. coli strains 6M and C6O were capable of invading HeLa cells (Fig. 4), as observed by microscopy and confirmed by the gentamicin protection (invasion) assay.
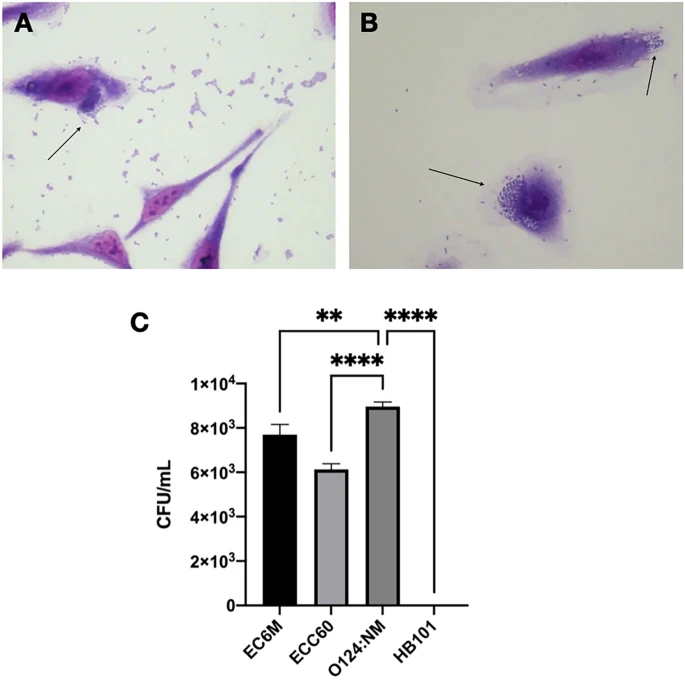
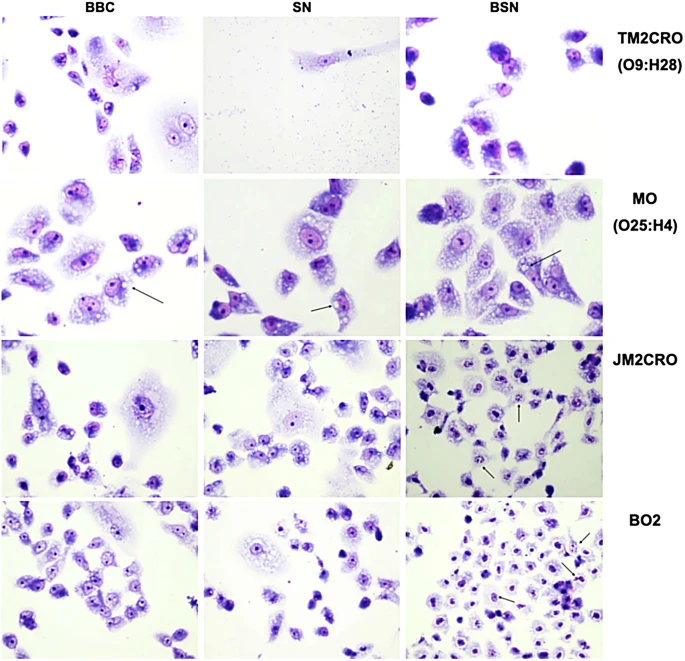
After the MO, TM2CRO, and JM2CRO strains were visually confirmed to be hazardous to HeLa cells, cytotoxic experiments on Vero cells were also carried out (Fig. 5). Surprisingly, all strains were hazardous to Vero cells when treated with live bacterial cells. Vero cells were cultured with dead bacteria to see if this was produced by cell–cell contact, but no effect was detected. To see if secreted proteins were involved in toxicity, filtered supernatant from various bacterial strains was cultured with Vero cells, and only the TM2CRO strain caused cell damage. A boiling, filtered supernatant was added to the cells to see if any produced toxins were thermostable. Because no effect on cells was seen after this treatment, this data suggested that E. E. coli TM2CRO expresses a thermolabile toxin. Surprisingly, a vacuolating effect was detected in all MO treatments, suggesting that this strain expresses a thermostable secreted toxin. In the case of the K. Boiling supernatants of JM2CRO and BO2 induced nuclear damage in Vero cells, showing that these strains release a thermostable toxin.
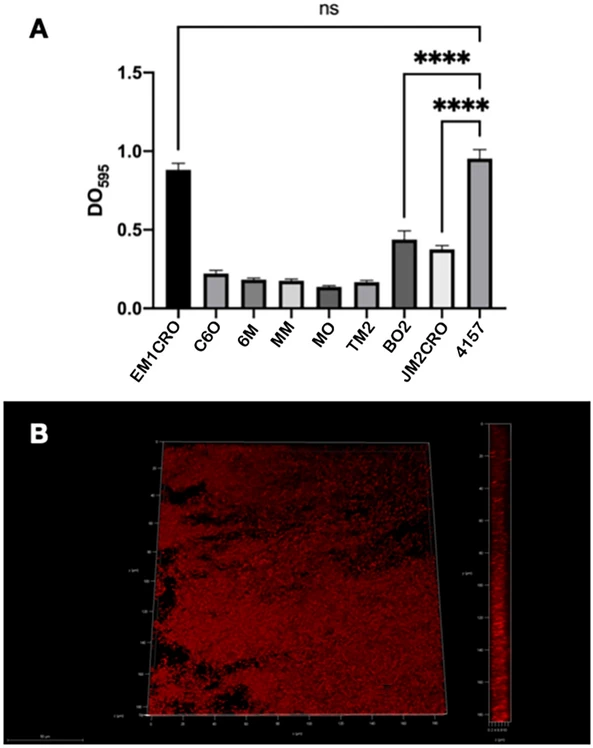
All strains tested in this study were able to produce biofilms, albeit the majority were thought to develop weak or moderate biofilms under the testing conditions (Fig. 6a). The EM1CRO strain, on the other hand, was an anomaly since it generated a biofilm as robust as E. coli 4157, which is regarded as a highly biofilm-forming control strain (Fig. 6b).
Discussion
The prevalence of ESBL and/or pAmpC-positive K. pneumoniae and E. coli strains contaminating bivalves in 6 out of 14 studied geographical points shows substantial contamination of this geographical region’s maritime environment with important priority bacteria. Indeed, adjacent cities have a history of marine pollution as a result of a lack of sewage treatment and sanitation facilities, with the existence of critical priority bacteria in their coastal waters and additional contamination of wild-caught fishes recently documented21,22,23,24,25. Because marine bivalves are filter-feeding creatures that remove a substantial quantity of suspended material from the water, the presence of human fecal pollution in these coastal waters can support the formation of bioaccumulation of such infections.
While E. coli is a genetically diverse species capable of causing both diarrheal disorders and MDR extra-intestinal infections, K. pneumoniae has been identified as a major cause of antimicrobial-resistant healthcare-associated infections. Furthermore, the global spread of these species has been linked to well-established evolutionary harmful lineages like K. pneumoniae ST307 and E. coli ST131, which frequently exhibit an MDR phenotype and a combination of virulence genes26, 27.
Worryingly, we discovered transnational CTX-M-27-producing E. coli ST131 and CTX-M-15-producing K. pneumoniae ST307 clones in oyster and mussel samples, respectively, in our analysis. This should be regarded as an epidemiological warning, as multiple investigations have revealed the rapid dissemination of these high-risk MDR clones at the human-animal interface, with several outbreaks occurring worldwide28, 29, 30.
The dissemination of virulence and MDR profiles is directly related to the presence of mobile genetic elements, in which plasmids are thought to play an important role. A direct relationship has been shown between high-risk strains and the presence of IncF plasmids, particularly those bearing FIA and FII replicon types, which were found in the majority of bacteria recovered in this study.
Currently, E. coli ST131 is the most common ExPEC clone circulating globally, consisting of numerous strains with unique resistance profiles33, suggesting that its emergence is involved in many events associated with horizontal transfer of antimicrobial resistance genes. Indeed, each ST131 subclone is linked to a different allele of the type 1 fimbrial adhesin gene (fimH), with H30 being the most common. It is worth noting that E. coli ST131-fimH30 has been linked to urinary tract infections all across the world, and it is commonly related to fluoroquinolone resistance34. On the other hand, subclone H22 has been linked to foodborne infections35. Although the E. coli ST131 found in the study belongs to subclone H30, it may provide a risk for foodborne infections because it has previously been isolated from food-producing animals, particularly retail chicken, 36, 37. Surprisingly, the majority of the viruses in subclone H30 are serotype O25:H4. The E. coli ST131-fimH30 (MO strain) was identified as an H4 type flagellum in this investigation, which possesses properties that aid in host colonization processes and is also capable of inducing greater induction of the cytokine IL-10, which contributes to its fitness in the urinary tract38.