Healthy eating and exercise are examples of lifestyle choices that can have a significant impact on one’s overall health. However, a person’s genetic makeup predominately determines their risk of developing cancer.
For the purpose of creating new cells, our bodies replicate our genes constantly. The phenomenon geneticists refer to as mutation occurs when there are sporadic errors in those copies. Some of these errors can change how a gene is copied, alter proteins, fuse genes, and affect how much a gene is expressed, all of which can affect a person’s risk of getting cancer. By creating predictive models for tumor activity, researchers can better comprehend the effects of mutations.
Assistant professor of biomedical engineering Christopher Plaisier at the Ira A. Fulton Schools of Engineering at Arizona State University is creating a software program called OncoMerge that uses genetic data to advance cancer modeling technology.
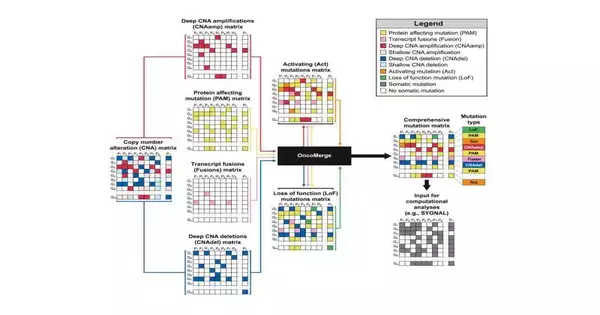
OncoMerge is a platform that recognizes abnormal gene fusions as well as mutations that impact protein expression and the frequency of gene duplication. Following that, the software examines the network underlying the mutations to identify relationships and create a model to forecast subsequent changes brought on by the mutations.
According to Plaisier, who is also an associate faculty member at the ASU Biodesign Center for Biocomputing, Security, and Society, “we are able to look at the gene expression patterns using correlation.”. Then, we can observe what is being repressed or activated, enabling us to examine the deeper functions underlying that.
Since his postdoctoral work, when he first realized the need for a platform that could process the network behind mutations, Plaisier has been considering the concept of OncoMerge. His expertise as a cancer biologist, computational biologist, and human geneticist are all combined into one project in this effort.
In his most recent work, he developed a database that uses genetic data to examine linked activity within networks in order to address challenges in the detection of gene mutations. Today, the findings were published in the journal Cell Reports Methods.
Data-driven research.
Numerous health care applications may result from studying genetics, but understanding cancer is one of their most important uses. Plaisier is improving prediction models that can provide insight into particular cancer environments due to the significant variation among cancer types.
His team tested the feedback networks of specific genomic regions to establish an abnormal regulation of networks before designing OncoMerge. This deliberate cultivation revealed how the tumor controlled its surroundings to survive.
In order to verify the effectiveness of the team’s methods, OncoMerge has been used on over 9,000 patient tumors. The results show that integrating mutation data did increase the precision of predictions for linked behavior among genes.
Plaisier anticipates using his software to analyze cancer mutation pipelines and eventually enhance precision and process genetic data in individual cells.
Educational data science
Plaisier wants to advance data science education along with enhancing cancer modeling methods. In addition to actively advocating for the inclusion of more introductory coding courses across majors to better acquaint students with coding techniques, he would like to develop a data science elective course for biomedical engineering students.
Graduate student in biological design, Sierra Wilferd, in Plaisier’s lab, values the interdisciplinary approach.
The interaction between cancer biology and bioinformatics, according to Wilferd, “has taught me the importance of applying lab skills and computational skills to ask and answer interesting questions.”
Since biological systems in humans are incredibly complex, understanding and collecting biological data are essential for preventing and treating disease. The OncoMerge software from Plaisier may play a significant role in the genomics data pipeline for studies of patient tumors and have a major impact on all cancer genomics studies.
More information: Christopher L Plaisier et al, Systematic integration of protein affecting mutations, gene fusions, and copy number alterations into a comprehensive somatic mutational profile, Cell Reports Methods (2023). DOI: 10.1016/j.crmeth.2023.100442. www.cell.com/cell-reports-meth … 2667-2375(23)00057-7