Cyclosporine is one of the most widely recognized and successful immunosuppressant drugs used to deal with ongoing illnesses like joint pain and psoriasis, yet it comes with a gamble of serious secondary effects. Researchers imagine that might be on the grounds that the medication extensively targets cyclophilins, a group of 17 administrative proteins that play various parts in advancing cell well-being. Although every individual cyclophilin subtype plays a one-of-a-kind part, numerous ongoing immunosuppressive medications focus on the whole family, implying that significant obscure pathways might be incidentally switched off or generally modified.
The issue is complicated by the way that the dynamic site where particles bind is practically indistinguishable across each of the 17 cyclophilins, making it extremely difficult for drugmakers to target explicit subtypes. In a paper distributed today in Nature Compound Science, researchers in the lab of Expansive Establishment Center Part David Liu, who is likewise the overseer of the Merkin Organization of Groundbreaking Advancements in Medical Care at Wide, as a team with the labs of Markus Seeliger at SUNY Stony Stream and Foundation Part Vamsi Mootha at Massachusetts General Emergency Clinic, have proposed another arrangement.
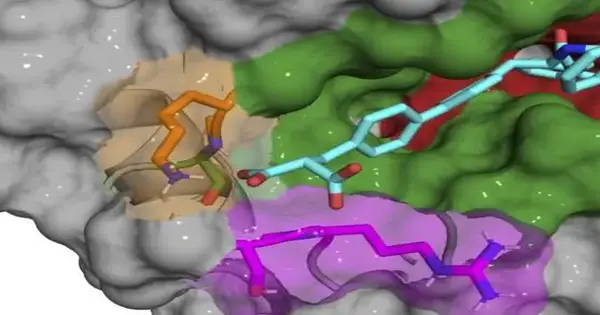
As opposed to focusing on the dynamic site of cyclophilin proteins, specialists in Liu’s lab depict a cycle that tracks down and intensifies that tight spot to the “exo site,” a little pocket close to the dynamic site that changes in size and shape across various cyclophilins. Involving secluded proteins in a test tube, the group found a few mixtures that solely tie and repress Cyclophilin D (CypD), a protein engaged in the opening and shutting of mitochondrial pores. They likewise applied comparable standards to find remarkable, specific inhibitors for Cyclophilin E (CypE). The creators say that their review lays the groundwork for researchers to create extra subtype-particular cyclophilin inhibitors, some of which might be valuable as instruments for science or as leads for helpful turns of events.
“It’s a new binding method that takes advantage of an untapped pocket. It serves as a model for future selective cyclophilin inhibitor development.”
Author Alex Peterson, now a postdoctoral fellow at the Scripps Research Institute
Another limiting mode exploits a pocket that individuals haven’t completely investigated at this point, “lead creator” Alex Peterson, presently a postdoctoral individual at the Scripps Exploration Foundation, who drove the task as an alumni understudy in Liu’s lab, said. “It’s sort of a plan for how individuals can plan specific cyclophilin inhibitors proceeding.”
Utilizing new and old advances in
CypD controls the mitochondrial penetrability change pore (mPTP), little pores situated on the inward surface of the mitochondria (broadly known as the force to be reckoned with of the cell). At the point when CypD distinguishes oxidative pressure or high calcium levels, it races to open the mPTP, permitting water and different particles to rush all through the mitochondria.
This opening of the mitochondrial conduits can turn into an issue with infections like ischemia reperfusion injury, diabetes, neurodegenerative problems, liver illnesses, and that’s only the tip of the iceberg. Since these circumstances can cause strangely elevated degrees of oxidative pressure, CypD holds the mitochondrial pores open for longer than expected, causing mitochondrial brokenness, cracking, and cell passing. It’s been felt that sedatives that lull and restrain CypD’s response to high oxidative pressure may be utilized to treat a large group of illnesses.
To find intensifies that solely tie to CypD, the group went to DNA-encoded little particle libraries, an innovation created a while back as one of the principal undertakings in Liu’s then-new lab. Scientists can utilize the libraries, which are loaded up with a huge number of engineered compounds connected to one-of-a-kind DNA standardized tags, to check for particles that tightly spot wanted proteins. By blending disengaged CypD proteins and an assortment of 256,000 one-of-a-kind DNA-encoded intensifies in a test tube, the group distinguished many promising mixtures.
A large portion of the underlying mixtures are actually bound in and around the dynamic site, repressing numerous cyclophilin subtypes, so the group step by step rolled out little substance improvements to their mixtures to make them exceptional to CypD. When they found that the exo site was the way to create subtype-explicit inhibitors, they had the option to plan a couple of mixtures that would powerfully repress CypD while insignificantly influencing other cyclophilins. The X-beam co-precious stone designs of the CypD protein and the inhibitors during improvement gave the group an in-depth take a gander at the exact place where their particles were restricted.
The scientists then treated detached mitochondria with their two driving mixtures and saw that they were powerful in dialing back CypD’s opening of mitochondrial pores. The identical representations of their mixtures, which don’t hinder CypD, didn’t show movement in mitochondria. To demonstrate that their prosperity was definitely not a disengaged occurrence, they rehashed the procedure for CypE, a cyclophilin liable for directing mRNA handling. Indeed, they cultivated a compound that was solely designated for it and left the excess 16 cyclophilins unphased.
The group believes that their findings will eventually help compound scholars and drugmakers develop better and more unambiguous cyclophilin-focusing drugs.They even gave future researchers an advantage—in light of the fact that the CypD-focusing on intensifies the battle to enter human cells all alone, the group changed them by adding ester subsidiaries that really sidestep the plasma layer and convey into mitochondria.
“Our cooperation in the end permitted us to overcome this well-established issue: how would you specifically restrain only one cyclophilin subtype out of 17?” said Liu, who is likewise a Howard Hughes Clinical Organization specialist. “Later on, particles that come from utilizing our technique will, I trust, end up being valuable both for fundamental science and possibly for therapeutics.”
More information: Alexander A. Peterson et al, Discovery and molecular basis of subtype-selective cyclophilin inhibitors, Nature Chemical Biology (2022). DOI: 10.1038/s41589-022-01116-1
Journal information: Nature Chemical Biology