An exploration group from the College of Hong Kong (HKU) led by Teacher Xiang David Li from the Branch of Science has, in a joint effort with Dr. Yuanliang Zhai from the HKU School of Natural Sciences and Dr. Jason Wing Hon Wong and Dr. Xiucong Bao from the HKU School of Biomedical Sciences, made a critical leap in understanding how hereditary data encoded in our DNA is perused and why blunders in perusing such data can frequently prompt formative imperfections or diseases. The discoveries were recently published in Science.
Each kind of cell in the human body (with certain exemptions) contains the very same DNA sequence, known as a “quality.” Hence, to make a particular cell type (e.g., an immature microorganism or a neuron), every cell needs to painstakingly pick which qualities to communicate. This essential cycle is directed by different adjustments of histone proteins, which were recently considered simple spools for bundling DNA in the core of our cells.
We currently realize that these histone changes are labels or checks on chromatin that have the same capability as expert switches for the guideline of qualities—tthey figure out which sets of qualities in a cell ought to be “on” or “off” with impeccable timing and for the right span. Dysregulation of this central cycle underlies numerous extreme human illnesses, like malignant growths.
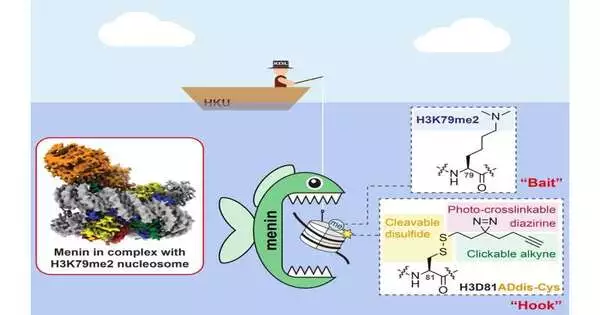
“The secret to success is not only the bait, but also a specifically engineered hook with a light-activated chemical group to grab the readers when exposed to UV light,”
Professor Xiang David Li from the Department of Chemistry
Various histone marks act as cell signs to control various chromatin-related apparatuses that direct quality articulation, DNA replication, and damage repair.One of the difficulties in chromatin science is the means by which specific histone marks are deciphered to accomplish their organic capability. To respond to this inquiry, it is fundamental to find the perusers, a class of proteins that perceive explicit histone checks and decipher them by turning the statement of qualities up or down as needs be.
However, the readers of numerous histone marks are currently unknown, limiting our ability to figure out their roles in quality guidelines.A well-established interest of Teacher Li’s lab is the improvement of novel synthetic ways to deal with recognized histone perusers that may be hard to track down utilizing customary natural strategies.
One such strategy utilizes a peptide containing a histone mark (i.e., a little section of histone protein) that goes about as the snare to look for perusers that perceive the imprint. “The way forward isn’t just the lure, but additionally an exceptionally planned snare that is furnished with a light-initiated substance gathering to catch the perusers upon openness to UV light,” Teacher Li explained.
In this review, the group zeroed in on a methylation mark at histone H3 lysine 79 (H3K79me2). In human cells, this imprint is tracked down through effectively communicated qualities. Deficiency of H3K79me2 in mammalian undeveloped organisms can prompt numerous formative irregularities, including disabled development, heart widening, and demise. Then again, H3K79me2 has been found at unusually undeniable levels and in some unacceptable spots (e.g., disease-advancing qualities) in different malignant growths like youth leukemia.
Regardless of its natural importance in quality guidelines, the system of how this imprint is “decoded” is hazy, as the perusers of H3K79me2 have not been found since its disclosure quite a while back. As a matter of fact, throughout the long term, numerous labs have attempted different ways to deal with the search for these perusers. “It is an incredible test to recognize H3K79me2 perusers, even with our recently evolved novel substance,” said Teacher Li.
There are two significant obstacles to overcome. To begin with, “pursuing” the imprints might include the actual imprint as well as the entire histone and, surprisingly, the histone-DNA complex called a nucleosome. Finally, perceiving H3K79me2 by its users may necessitate a local nucleosome–or chromatin–setting.Second, the cooperation between the perusers and H3K79me2 can be feeble or even transient, and in this manner, it is effortlessly lost during the fishing system.
“To catch H3K79me2 perusers, we should update our trap and snare,” said Li. Yet, it was not insignificant. Li’s lab spent over five years fostering their new device. Rather than utilizing a little section of the histone protein, they synthetically orchestrated an unblemished nucleosome with an updated tri-utilitarian snare and H3K79me2 as the lure. Using this new technology, the researchers successfully identified a protein called menin as the H3K79me2 peruser.
To comprehend how menin read the H3K79me2 mark, the group embraced a state-of-the art innovation called cryo-electron microscopy to envision the sub-atomic subtleties of this collaboration. “Untangling the nuances of how menin ties H3K79 methylation is critical to developing new medications to treat malignant growths associated with [dysregulated] H3K79me2,” said Teacher Li.
The spearheading work by Li and collaborators has advanced how we might interpret the key natural cycles of quality guidelines. These findings also pave the way for the development of novel therapeutic agents to treat human infections caused by high levels of H3K79 methylation.
More information: Jianwei Lin et al, Menin “reads” H3K79me2 mark in a nucleosomal context, Science (2023). DOI: 10.1126/science.adc9318
Journal information: Science