The history of human genetic variation can be seen in repetitive DNA sequences surrounding the centromere.
Scientists lied a little when they released the entire sequence of the human genome in 2003.
In actuality, nearly 20 years later, only approximately 8% of the genome had been fully sequenced, owing to highly repetitive DNA segments that are difficult to match with the rest of the genome.
However, a three-year-old team has finally filled in the gaps in the remaining DNA, giving scientists and medics the first complete, gap-free genome sequence.
The freshly completed genome, termed T2T-CHM13, is a significant improvement over the current reference genome, GRCh38, which is used by doctors and scientists to check for disease-linked mutations as well as to study the evolution of human genetic variation.
The new DNA sequences, among other things, disclose previously unknown details about the region surrounding the centromere, which is where chromosomes are grabbed and tugged apart when cells split, ensuring that each “daughter” cell acquires the correct number of chromosomes. Variation within this region could also reveal new information about how our ancestors evolved in Africa.
At the same time, “uncovering the whole sequence of these formerly missing parts of the genome informed us so much about how they’re organized,” Nicolas Altemose, a postdoctoral fellow at the University of California, Berkeley, and a co-author of four new articles describing the completed genome, said. Previously, we could only get a hazy picture of what was there; today it’s crystal clear down to single base pair resolution.
Altemose is the first author of a publication describing the base pair sequences surrounding the centromere. The journal Science will publish an article outlining how the sequencing was done on April 1, while Altemose’s centromere paper and four others discussing what the new sequences tell us will be summarized in the journal, with the complete papers available online. Four other papers, including one for which Altemose is co-first author, will be published in the journal Nature Methods on April 1.
The sequencing and analysis were carried out by the Telemere-to-Telomere Consortium, or T2T, a group of over 100 people named after the telomeres that cap the ends of all chromosomes. The gapless version of all 22 autosomes and the X chromosome created by the group has 3.055 billion base pairs (the building blocks of chromosomes and genes) and 19,969 protein-coding genes. The T2T researchers discovered around 2,000 new protein-coding genes, most of which were disabled, but 115 of which could still be produced. They also discovered around 2 million new variations in the human genome, 622 of which are identified in therapeutically significant genes.
In the future, when someone’s genome is sequenced, we’ll be able to identify all of the variants in their DNA and utilize that information to better direct their health care, said Adam Phillippy, one of T2leaders T’s and a senior investigator at the National Human Genome Research Institute (NHGRI). “It was like putting on a new pair of glasses when we finally finished the human genome sequencing.” We’re one step closer to comprehending what it all means now that we can see everything clearly. “
The centromere’s evolution
Additional DNA sequences in and around the centromere account for approximately 6.2 percent of the total genome, or nearly 190 million nucleotide base pairs.The majority of the newly inserted sequences are found near the telomeres at the ends of each chromosome, as well as in the areas surrounding ribosomal genes. The complete genome is made up of only four types of nucleotides, which code for the amino acids that make up proteins in groups of three. The main focus of Altemose’s study is to identify and investigate sites on the chromosomes where proteins interact with DNA.
“DNA is nothing without proteins,” said Altemose, who acquired a Ph.D. in bioengineering from UC Berkeley and UC San Francisco in 2021, following a D.Phil. in statistics from Oxford University. If proteins aren’t around to organize it, control it, repair it when it’s damaged, and reproduce it, DNA is a collection of instructions with no one to interpret them. Protein-DNA interactions are where all of the action for genome regulation takes place, and being able to map where specific proteins bind to the genome is critical for understanding their function.
Altemose and his team employed new tools to determine the location within the centromere where a large protein complex called the kinetochore tightly grips the chromosome so that other machineries inside the nucleus may tug chromosomal pairs apart after the T2T consortium reads the missing DNA.
“When this goes wrong, you end up with missegregated chromosomes, which causes a slew of issues,” he explained. “Chromosomal abnormalities can lead to spontaneous miscarriage or congenital illnesses if this happens during meiosis.” If it happens in somatic cells, it can lead to cancer, which is defined as “cells with significant misregulation.”
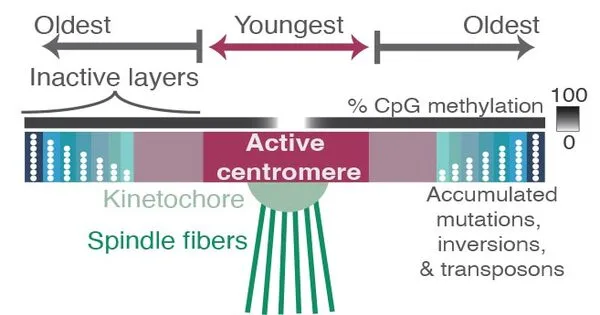
They discovered layers of new sequences on top of layers of older sequences in and around the centromeres, as if fresh centromere regions have been laid down repeatedly to bind to the kinetochore through evolution. The older sections have more random mutations and deletions, indicating that the cell is no longer using them. The kinetochore attaches to younger sequences that are less varied and have less methylation. A methyl group is an epigenetic marker that has the ability to silence genes.
All of the layers in and around the centromere are made up of repeating DNA sequences based on a 171-base pair unit, which is roughly the length of DNA that wraps around a group of proteins to form a nucleosome, keeping the DNA packaged and compact. These 171 base pair units are duplicated several times in tandem, forming a vast area of repetitive sequences around the centromere.
The T2T researchers only looked at one human genome, which came from a non-cancerous tumor known as a hydatidiform mole, which is essentially a human embryo that rejected maternal DNA and reproduced its father’s DNA instead. Embryos like these perish and turn into cancers. It was easier to sequence this mole because it had two identical copies of paternal DNA, both containing the father’s X chromosome, rather than distinct DNA from both mother and father.
According to Altemose, the researchers also revealed the whole sequence of a Y chromosome from a different source this week, which took nearly as long to construct as the rest of the genome combined. The results of this novel Y chromosomal sequence will be published in the near future.
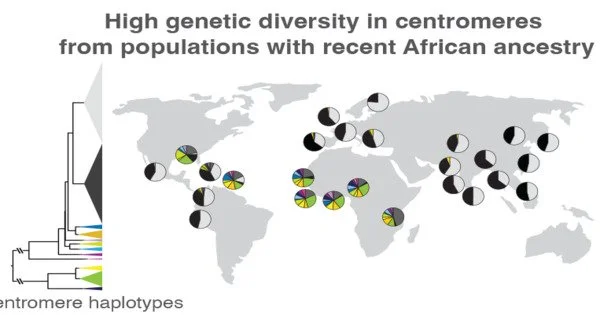
Altemose and his colleagues, which included UC Berkeley project scientist Sasha Langley, used the new reference genome as a scaffold to compare the centromeric DNA of 1,600 people from all over the world, revealing significant differences in the sequence and copy number of repetitive DNA around the centromere. Previous research has indicated that ancient humans only took a tiny sample of genetic variants with them when they moved out of Africa to the rest of the world. This pattern continues into centromeres, according to Altemose and his colleagues.
At least on chromosome X, people with recent ancestry outside of Africa have centromeres that fit into two big groups, whereas people with recent African heritage have the most fascinating variety, “Altemose added. “Given what we know about the remainder of the genome, this isn’t altogether surprising.” But it shows that if we want to investigate the intriguing variance in these centromeric areas, we need to make a concerted effort to sequence additional African genomes and perform comprehensive telomere-to-telomere sequence assembly.
He also mentioned that DNA sequences at the centromere could be utilized to trace human lineages back to our common ape ancestors.
The sequence degrades as it moves away from the active centromere, according to Altemose.Eventually, if you travel out to the farthest borders of this sea of repeated sequences, you start to see the ancient centromere that, possibly, our distant primate ancestors used to connect to the kinetochore.” It’s almost as though you’re looking at layers of fossils.
A game-changer is long-read sequencing.
The success of the T2T is attributed to improved techniques for sequencing long lengths of DNA at once, which aid in establishing the order of highly repetitive DNA segments. PacBio’s HiFi sequencing, for example, can read lengths of more than 20,000 base pairs with great precision. On the other hand, Oxford Nanopore Technologies Ltd. has created technology that can read up to several million base pairs in succession, though with less precision. For example, Illumina Inc.’s so-called next-generation sequencing is restricted to hundreds of base pairs.
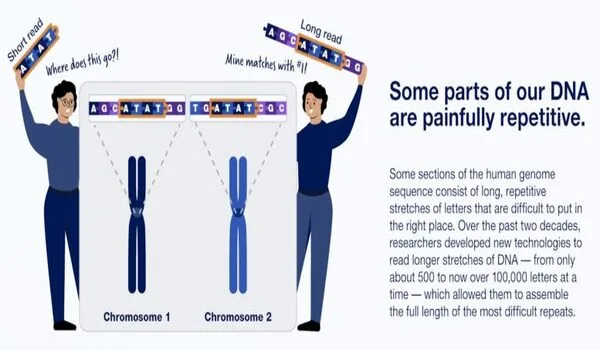
“These new long-read DNA sequencing technologies are simply wonderful; they’re such game changers, not only for this repetitive DNA world, but also because they allow you to sequence single lengthy molecules of DNA,” said Altemose. “With short-read sequencing technologies, you can start asking questions at a level of resolution that wasn’t conceivable previously.”
“These new long-read DNA sequencing technologies are simply wonderful; they’re such game changers, not only for this repetitive DNA world, but also because they allow you to sequence single lengthy molecules of DNA, With short-read sequencing technologies, you can start asking questions at a level of resolution that wasn’t conceivable previously.”
said Altemose.
Altemose wants to go deeper into the centromeric areas, utilizing a new technology he and Stanford colleagues devised to find protein-binding locations on the chromosome, similar to how the kinetochore connects to the centromere. Long-read sequencing technology is used in this procedure as well. In a report published this week in the journal Nature Methods, he and his colleagues detailed the process, dubbed Directed Methylation with Long-read Sequencing (DiMeLo-seq).
In the meantime, the T2T consortium is collaborating with the Human PanGenome Reference Consortium to create a reference genome that represents the whole human race.
“Instead of having one reference from one human or one hydatidiform mole, which isn’t even a real human individual,” Altemose added, “we should have a reference that represents everyone.” “There are a variety of approaches to achieving that goal. But first, we need to understand what that variation looks like, which requires a large number of high-quality individual genome sequences.
Postdoctoral scholarships supported his work on centromeric regions, which he described as “a love endeavor.” Karen Miga of UC Santa Cruz, Evan Eichler of the University of Washington, and Adam Phillippy of the National Human Genome Research Institute, which supplied much of the financing, led the T2T research. The other UC Berkeley co-authors of the centromere paper are Aaron Streets, associate professor of bioengineering; Abby Dernburg and Gary Karpen, professors of molecular and cell biology; project scientist Sasha Langley; and former postdoctoral fellow Gina Caldas.
Reference: “Complete genomic and epigenetic maps of human centromeres” by Nicolas Altemose, Glennis A. Logsdon, Andrey V. Bzikadze, Pragya Sidhwani, Sasha A. Langley, Gina V. Caldas, Savannah J. Hoyt, Lev Uralsky, Fedor D. Ryabov, Colin J. Shew, Michael E. G. Sauria, Matthew Borchers, Ariel Gershman, Alla Mikheenko, Valery A. Shepelev, Tatiana Dvorkina, Olga Kunyavskaya, Mitchell R. Vollger, Arang Rhie, Ann M. McCartney, Mobin Asri, Ryan Lorig-Roach, Kishwar Shafin, Julian K. Lucas, Sergey Aganezov, Daniel Olson, Leonardo Gomes de Lima, Tamara Potapova, Gabrielle A. Hartley, Marina Haukness, Peter Kerpedjiev, Fedor Gusev, Kristof Tigyi, Shelise Brooks, Alice Young, Sergey Nurk, Sergey Koren, Sofie R. Salama, Benedict Paten, Evgeny I. Rogaev, Aaron Streets, Gary H. Karpen, Abby F. Dernburg, Beth A. Sullivan, Aaron F. Straight, Travis J. Wheeler, Jennifer L. Gerton, Evan E. Eichler, Adam M. Phillippy, Winston Timp, Megan Y. Dennis, Rachel J. O’Neill, Justin M. Zook, Michael C. Schatz, Pavel A. Pevzner, Mark Diekhans, Charles H. Langley, Ivan A. Alexandrov and Karen H. Miga, 1 April 2022, Science.





